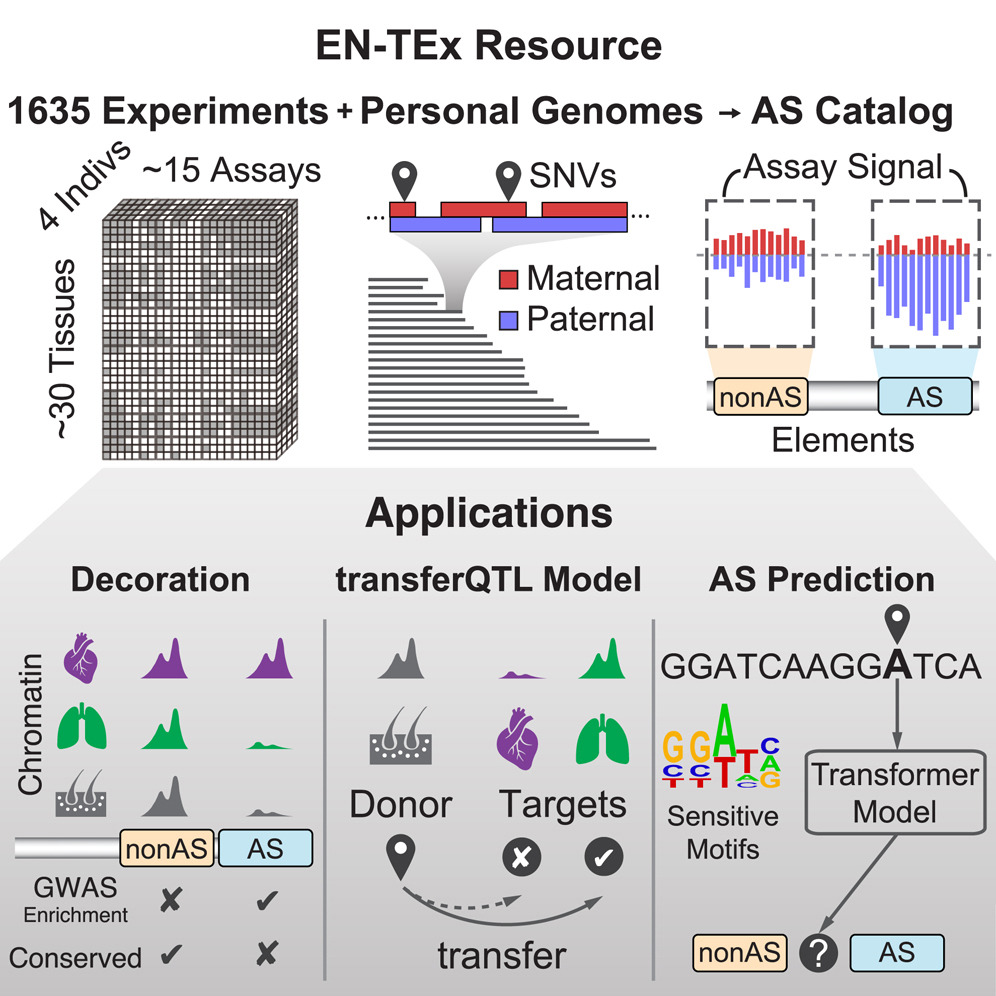
The EN-TEx resource of multi-tissue personal epigenomes & variant-impact models
Joel Rozowsky, Jiahao Gao, Beatrice Borsari, Yucheng T. Yang, Timur Galeev, Gamze Gürsoy, Charles B. Epstein, Kun Xiong, Jinrui Xu, Tianxiao Li, Jason Liu, Keyang Yu, Ana Berthel, Zhanlin Chen, Fabio Navarro, Maxwell S. Sun, James Wright, Justin Chang, Christopher J.F. Cameron, Noam Shoresh, Elizabeth Gaskell, Jorg Drenkow, Jessika Adrian, Sergey Aganezov, François Aguet, Gabriela Balderrama-Gutierrez, Samridhi Banskota, Guillermo Barreto Corona, Sora Chee, Surya B. Chhetri, Gabriel Conte Cortez Martins, Cassidy Danyko, Carrie A. Davis, Daniel Farid, Nina P. Farrell, Idan Gabdank, Yoel Gofin, David U. Gorkin, Mengting Gu, Vivian Hecht, Benjamin C. Hitz, Robbyn Issner, Yunzhe Jiang, Melanie Kirsche, Xiangmeng Kong, Bonita R. Lam, Shantao Li, Bian Li, Xiqi Li, Khine Zin Lin, Ruibang Luo, Mark Mackiewicz, Ran Meng, Jill E. Moore, Jonathan Mudge, Nicholas Nelson, Chad Nusbaum, Ioann Popov, Henry E. Pratt, Yunjiang Qiu, Srividya Ramakrishnan, Joe Raymond, Leonidas Salichos, Alexandra Scavelli, Jacob M. Schreiber, Fritz J. Sedlazeck, Lei Hoon See, Rachel M. Sherman, Xu Shi, Minyi Shi, Cricket Alicia Sloan, J Seth Strattan, Zhen Tan, Forrest Y. Tanaka, Anna Vlasova, Jun Wang, Jonathan Werner, Brian Williams, Min Xu, Chengfei Yan, Lu Yu, Christopher Zaleski, Jing Zhang, Kristin Ardlie, J Michael Cherry, Eric M. Mendenhall, William S. Noble, Zhiping Weng, Morgan E. Levine, Alexander Dobin, Barbara Wold, Ali Mortazavi, Bing Ren, Jesse Gillis, Richard M. Myers, Michael P. Snyder, Jyoti Choudhary, Aleksandar Milosavljevic, Michael C. Schatz, Bradley E. Bernstein, Roderic Guigó, Thomas R. Gingeras, and Mark Gerstein
Cell, Mar 2023
Understanding how genetic variants impact molecular phenotypes is a key goal of functional genomics, currently hindered by reliance on a single haploid reference genome. Here, we present the EN-TEx resource of 1,635 open-access datasets from four donors (∼30 tissues × ∼15 assays). The datasets are mapped to matched, diploid genomes with long-read phasing and structural variants, instantiating a catalog of >1 million allele-specific loci. These loci exhibit coordinated activity along haplotypes and are less conserved than corresponding, non-allele-specific ones. Surprisingly, a deep-learning transformer model can predict the allele-specific activity based only on local nucleotide-sequence context, highlighting the importance of transcription-factor-binding motifs particularly sensitive to variants. Furthermore, combining EN-TEx with existing genome annotations reveals strong associations between allele-specific and GWAS loci. It also enables models for transferring known eQTLs to difficult-to-profile tissues (e.g., from skin to heart). Overall, EN-TEx provides rich data and generalizable models for more accurate personal functional genomics.
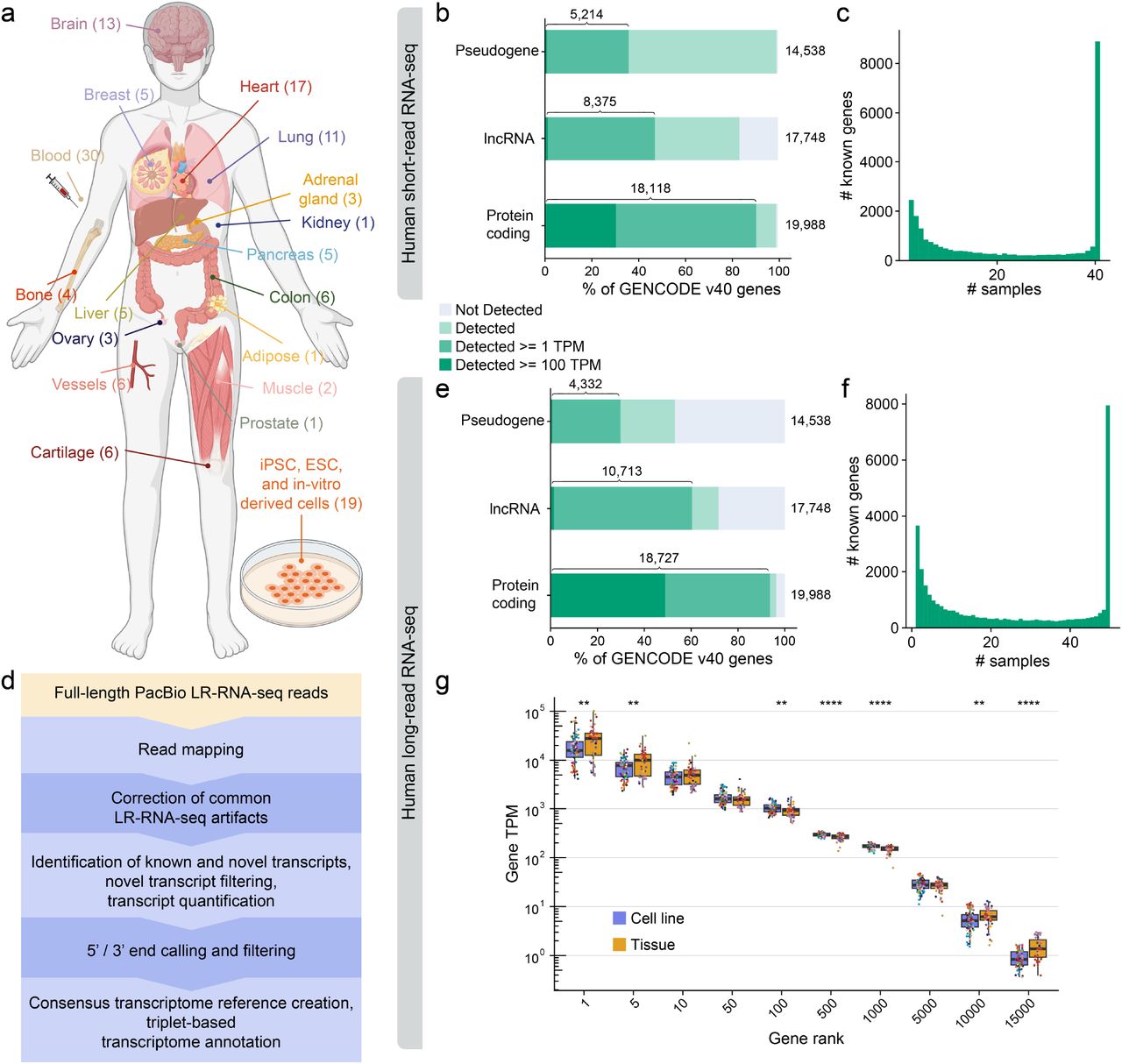 The ENCODE4 long-read RNA-seq collection reveals distinct classes of transcript structure diversitybioRxiv, 2023
The ENCODE4 long-read RNA-seq collection reveals distinct classes of transcript structure diversitybioRxiv, 2023